Researchers create ‘lipidomic map’ of tuberculosis bacteria, offering insights into immunology
An international team of scientists has developed a method to detect thousands of lipid molecules that are presented to the human immune system. This information can be used for the development of vaccines or anti-microbial treatments for tuberculosis.
FSE Science Newsroom |René Fransen and Harvard University
The study, co-led by D. Branch Moody, MD, from the Brigham and Women’s Hospital, (Boston, USA), and J. Rossjohn from Monash University (Australia), represents a collaboration among researchers from the USA, United Kingdom, Australia, and the group of Adri Minnaard at the University of Groningen, the Netherlands. The results were published in the journal Cell on 18 September.
Until the 1990s, immunologists thought that only proteins could activate the immune system. Then, it was discovered that lipids could be captured by specialized molecules that present them to the immune system, thereby also activating an immune response. In the study presented in Cell, the team developed a new and sensitive method to detect more than 2,000 lipids that are bound to CD1 antigen-presenting molecules, which present antigens to the human immune system.
(Story continues below the illustration)
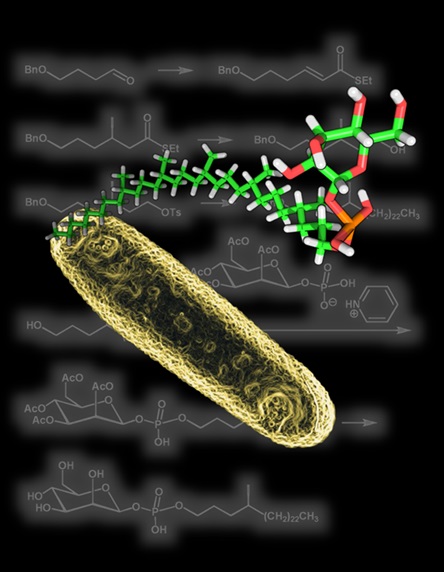
Producing complex lipids
Because lipids are not encoded by genes and form into membranes, they have entirely different functions and positions within the cell. The ability to measure many lipid antigens at any one time will allow future researchers to cross-check any disease-related lipid of interest against the list of candidate lipid antigens from this map and potentially make connections to diseases.
The Minnaard lab has specialized in producing the complex lipids
Importantly, Mycobacterium tuberculosis, the bacterium causing TB, produces many lipids that bind to CD1 antigen-presenting molecules. For this study, most of these lipids have been chemically prepared by the Minnaard group. ‘For a number of reasons, it is very difficult to isolate lipids from this bacterium’, says Minnaard. Therefore, the only way to obtain these molecules for scientific studies is by synthesizing them, and this process takes years to develop for each molecule. The Minnaard lab has specialized in producing the complex lipids from M. tuberculosis.
The structural biology part of the work, undertaken in Rossjohn's lab, shows how lipids fit inside proteins using size-based mechanisms. Combined, the structures of the proteins and lipids provide rules about the size, shape, and chemical content of the kinds of lipids that can bind CD1 and cause a T-cell response—either activation or deactivation. It is the latest in a series of studies that date back to the 1990s, when 'scientists from Brigham and Women's Hospital discovered that T cells can recognize lipid antigens.
(Story continues below the illustration)

Vaccine
‘I applaud the tenacity of the researchers in building the critical mass of technology needed to develop this multi-stage system that allows large numbers of lipids to be identified, solved individually, and then grouped into patterns,' says Moody. ‘This multidisciplinary effort involved biophysical techniques that are related to mass spectrometry and biological techniques related to lipid chemistry. The lipids informed immunological outputs, and the mode of lipid recognition was proven using X-ray crystallography.’
Identifying how lipids from Mycobacteria activate the immune system is important, explains Minnaard: ‘With a vaccine, you want to stimulate those cells of the immune system that respond to lipids from this bacteria. Now that we know how different lipids are presented to the immune system, we can check if potential vaccines that are based on lipids produce the most effective immune response.’ Also, some lipids reduce the response by the immune system. These could be the target of a treatment, for example against the Mycobacterium that causes leprosy.
(Story continues below the illustration)
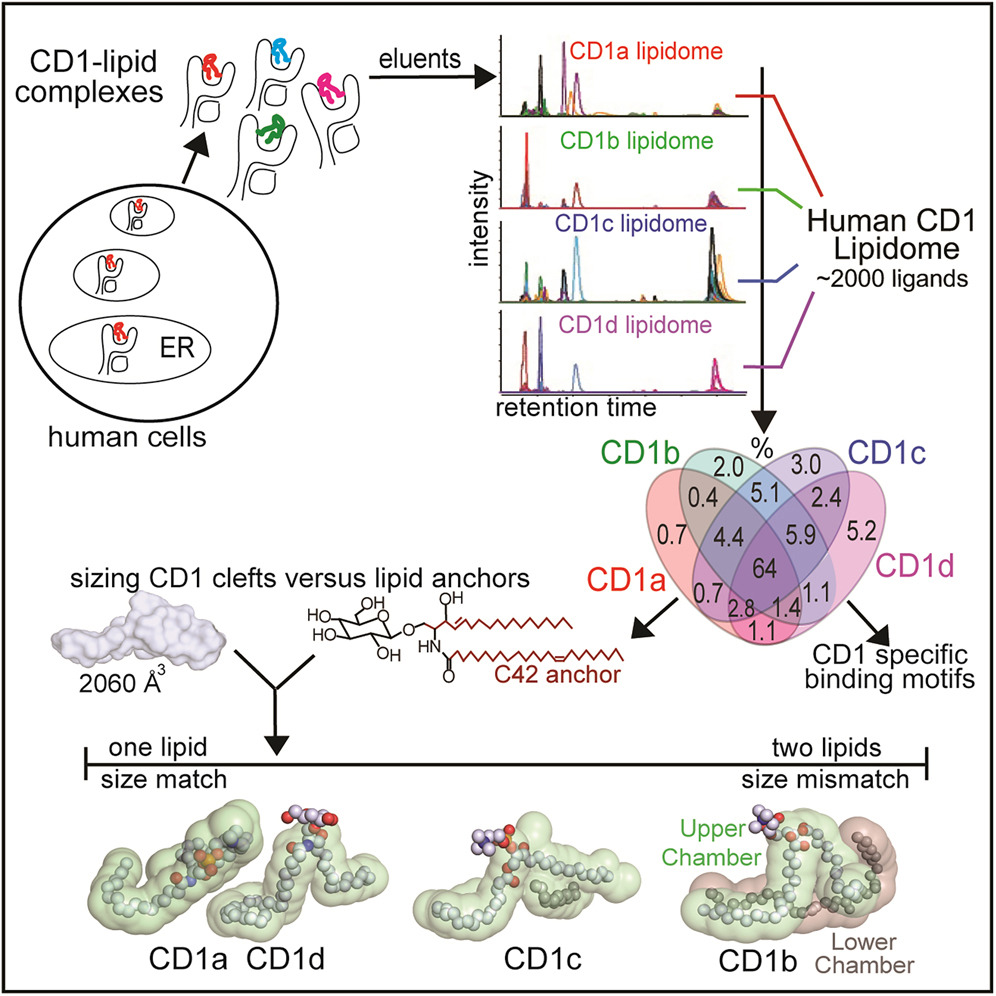
Reference: Shouxiong Huang, Adam Shahine, Tan-Yun Cheng, Yi-Ling Chen, Soo Weei Ng, Rachel Farquhar, Stephanie Gras, Clare S. Hardman, John D. Altman, Nabil Tahiri, Adriaan J. Minnaard, Graham S. Ogg, Jacob A. Mayfield, Jamie Rossjohn, and D. Branch Moody: CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms . Cell, 18 September 2023
| Last modified: | 27 June 2024 3.46 p.m. |
More news
-
21 November 2024
Dutch Research Agenda funding for research to improve climate policy
Michele Cucuzzella and Ming Cao are partners in the research programme ‘Behavioural Insights for Climate Policy’
-
13 November 2024
Can we live on our planet without destroying it?
How much land, water, and other resources does our lifestyle require? And how can we adapt this lifestyle to stay within the limits of what the Earth can give?
-
13 November 2024
Emergentie-onderzoek in de kosmologie ontvangt NWA-ORC-subsidie
Emergentie in de kosmologie - Het doel van het onderzoek is oa te begrijpen hoe ruimte, tijd, zwaartekracht en het universum uit bijna niets lijken te ontstaan. Meer informatie hierover in het nieuwsbericht.

