Nanopores suitable for single-molecule identification and sequencing of complete proteins
Proteins are vital to our bodies. As different tissues often express different proteins in different concentrations, sometimes with modifications, it is important to be able to study single proteins in detail. Scientists at the University of Groningen have published two papers in short succession, in Nature Chemistry and in Nature Communications, in which they describe how to construct a proteosome on top of a nanopore, which can be used for protein identification at a single-molecule level by fingerprinting and – in the future – by sequencing.
These days, sequencing a person’s DNA is fast and cheap. But although the proteins in our bodies are produced based on our DNA, this does not mean that we know all about those proteins. ‘DNA is a static molecule,’ explains Giovanni Maglia, Professor of Chemical Biology at the Biomolecular Sciences and Biotechnology Institute of the University of Groningen. The protein that is produced based on the DNA code is much more dynamic. The concentration depends on the kind of tissue or even the type of cell you are looking at, while proteins can appear in different variants and may be modified after production.

Reaction vessel
The golden standard for protein analysis is mass spectrometry. ‘However, this requires a certain amount of protein; it is, therefore, not suitable for studying single proteins. Furthermore, mass spectrometry only identifies protein fragments (peptides) and much information on the heterogeneity of proteins is lost,’ says Maglia. He has ample experience in studying single proteins in nanopores. ‘This is a very promising technique to study entire proteins at the single-molecule level that will reveal much information about proteins that is now hidden. We propose two approaches: chop the protein into small parts and create a “single-molecule fingerprint” or directly sequence the entire protein.’
In two papers, published in Nature Communications on 4 October and in Nature Chemistry on 18 November, Maglia and his colleagues describe how a nanopore can recognize proteins from the peptide spectra and how a nanopore can be fitted with a reaction vessel in the form of a proteasome. The latter can chop the protein into smaller pieces that can be identified in the pore but that could – in the future – also be used to thread a single intact protein through the pore and sequence the amino acids directly.
Fingerprint
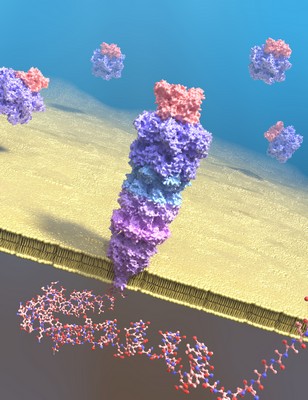
The Nature Communications paper shows that it is possible to identify a protein by measuring the fingerprint of predigested peptides using nanopores. ‘We used trypsin to create small uniform fragments and a low pH to allow detection of the fragments.’ In the Nature Chemistry article, the scientists describe how the complex proteasome-nanopore is constructed. ‘The proteasome is a complex system, created from 42 different pieces,’ explains Maglia. It contains sites to catch proteins, unfold them, and cut the protein into small peptides that are then passed through the pore. ‘The volume of the peptide produces a specific signal and this allows us to create a fingerprint with which we can identify the protein.’
Competition
The strength of the nanopore technique is that it identifies single proteins. The proteasome acts as the reaction vessel that prepares the protein for identification. ‘Our system works but it needs some improvements,’ says Maglia. The peptides pass through the pore too quickly to identify or sequence them. ‘If we use the “thread and read” option, we have to add a constriction to the pore, to below 1 nanometre.’ Once this is achieved, the intact polypeptides passing through the nanopore might be sequenced. ‘If we use the option to “chop and drop”, we will have to engineer the proteasome to cut proteins and engineer the nanopore to capture the majority of the peptide fragments.’
A great effort is being made – and there is much competition – to develop single-molecule protein sequencing. As these two articles were published, another approach appeared in the literature and was described independently by two groups: one also based in the Netherlands and another based in China. In that approach, small DNA molecules were attached to small protein fragments, which were then precisely moved across the nanopore using a helicase. ‘It is difficult to predict which approach will eventually prevail,’ says Maglia. ‘Their approach moves the protein fragments very precisely, while our approach has the advantage that entire proteins are addressed and it does not require DNA modifications.’
References:
Florian Lucas, Roderick Versloot, Liubov Yakovlieva, Marthe T. C. Walvoort & Giovanni Maglia: Protein identification by nanopore peptide profiling. Nature Communications 4 October 2021
Shengli Zhang, Gang Huang, Roderick Versloot, Bart Marlon Herwig Bruininks, Paulo Cesar Telles de Souza, Siewert-Jan Marrink, and Giovanni Maglia: Bottom-up fabrication of a proteasome–nanopore that unravels and processes single proteins. Nature Chemistry, 18 November 2021
| Last modified: | 18 November 2021 5.48 p.m. |
More news
-
10 June 2024
Swarming around a skyscraper
Every two weeks, UG Makers puts the spotlight on a researcher who has created something tangible, ranging from homemade measuring equipment for academic research to small or larger products that can change our daily lives. That is how UG...
-
21 May 2024
Results of 2024 University elections
The votes have been counted and the results of the University elections are in!

